1. configuration parameters
uDeviceX is configured via runtime parameters. These parameters are passed by the program via three mechanisms:
-
default parameters (priority 0)
-
file parameters
conf.cfg(optional) (priority 1) -
arguments
args(optional) (priority 2)
The program can be run as follows (for one node):
udx [conf.cfg] [args]When reading parameters, the program will select the one with highest priority. This makes it easy to overwrite the default parameters.
A file named conf.history.cfg is dumped once the program has read
all parameters. The file contains all parameters which have beed read
by udx.
1.1. Default parameters
We describe here the default parameters, contained in the file ${HOME}/.udx/default.cfg.
1.1.1. glb: global parameters
Global parameters
glb = {
restart = false (1)
kBT = 0.0444302 (2)
numdensity = 10 (3)
L = [8, 8, 8]
}| 1 | create initial conditions from restart files |
| 2 | temperature of the system (supposed uniform) |
| 3 | number density of solvent particles |
1.1.2. time
Time related parameters.
time = {
wall = 0.0 (1)
screenlog = false (2)
mbr_bb = 0.0
type = "const"
dt = 1e-4
end = 1.0
}| 1 | time of freezing step |
| 2 | dump the time step informations to the console |
1.1.3. flu
Describe the solvent parameters
flu = {
mass = 1.0 (1)
colors = false (2)
ids = false (3)
stresses = false (4)
recolor_freq = 0 (5)
push = true (6)
self = "pair_flu" (7)
}| 1 | mass of one particle |
| 2 | each particle has a different color if set to true |
| 3 | each particle has unique global index if set to true |
| 4 | compute solvent stresses if set to true |
| 5 | re-color the solvent according to membranes every x iterations;
0: deactivated; membranes must be active if set to non 0. |
| 6 | apply body forces on solvent if set to true |
| 7 | self interaction parameters (see below) |
The interaction parameters are described in a separate namespace.
In the flu.self case, the default is given by:
The pair interaction parameters have all the same structure, allowing to share namespaces for common interactions.
pair_flu = {
dpd = true (1)
adhesion = false (2)
lj = false (3)
a = [2.6666667, 2.6666667, 2.6666667] (4)
g = [8.0, 8.0, 8.0] (5)
spow = 0.25 (6)
}| 1 | activate DPD interactions |
| 2 | no repulsive Lennard-Jones potential |
| 3 | no adhesion interactions |
| 4 | repulsion parameters |
| 5 | dissipative parameters |
| 6 | envelope parameter for the random kernel |
Note that the parameters form an array.
These correspond to different color interactions.
In this case, a maximum of 2 colors is possible.
The parameters correspond to blue-blue, blue-red,red-red.
1.1.4. debug parameters
We list here default debug parameters
dflu = {
debug = false (1)
}
dbg = {
verbose = true (2)
dump = true (3)
pos = false (4)
pos_soft = false (5)
vel = false (6)
forces = false (7)
colors = false (8)
clist = false (9)
}| 1 | activate debug mode for solvent distribution |
| 2 | activate verbose for debug informations |
| 3 | activate dump informations for debug modes |
| 4 | check positions of solvent |
| 5 | check positions of solvent and objects |
| 6 | check velocities |
| 7 | check forces |
| 8 | check colors of solvent |
| 9 | check cell lists |
1.2. examples
We here list simple run cases of uDeviceX.
1.2.1. equilibration
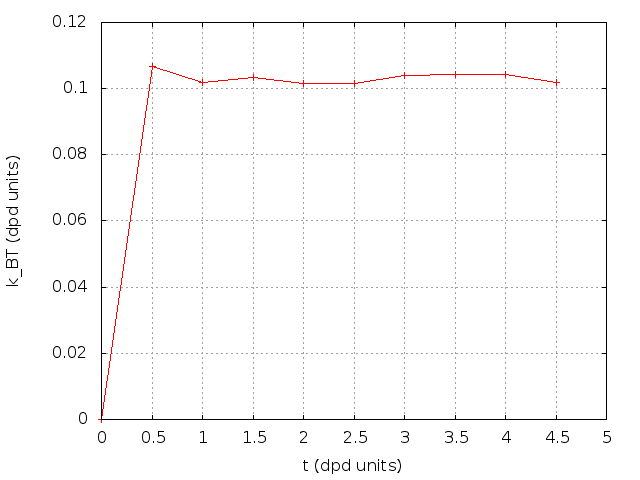
A simple solvent at rest equilibrating at given temperature. We can already see non default parameters appearing:
glb = {
L = [8, 8, 8] (1)
numdensity = 10
kBT = 0.1
}
time = {
type = "const" (2)
dt = 1e-3 (3)
end = 5.0 (4)
}| 1 | domain size |
| 2 | timestep mode: constant in this case |
| 3 | step size (in dpd time units) |
| 4 | simulation time (in dpd time units) |
The dpd parameters are set as follows:
flu = {
self = "flu_pair"
mass = 1.0
}
flu_pair = {
dpd = true (1)
lj = false (2)
a = [ 20.0 ] (3)
g = [ 8.0 ] (4)
spow = 0.25 (5)
adhesion = false
}| 1 | activate dpd interaction |
| 2 | no lj interactions (same as default) |
| 3 | repulsion parameter |
| 4 | dissipation parameter |
| 5 | "envelope" power for the random kernel |
Note that the parameters are inside an array. This is useful when we set different colors.
The temperature quickly equilibrates to the required value:
1.2.2. double_poiseuille
Reverse poiseuille flow in a periodic box. The setup looks really like the previous case equilibration, but we now add body forces:
bforce = {
type = "double_poiseuille" (1)
a = 1.0 (2)
}| 1 | type of body force |
| 2 | magnitude of the body force |
Note that we also need to activate the push option for solvent:
flu = {
mass = 1.0
self = "flu_pair"
push = true (1)
}
flu_pair = {
dpd = true
lj = false
adhesion = false
a = [ 20.0 ]
g = [ 8.0 ]
spow = 0.25
}| 1 | activate body force for solvent |
In order to visualize the flow, we choose here to dump field data. The data is sampled via the sampling module, activated when the field dump is active.
sampler = {
n_per_dump = 1000 (1)
grid_ref = [1, 1, 1] (2)
}
dump = {
freq_diag = 1.0 (3)
field = true (4)
freq_field = 2.0 (5)
}| 1 | number of samples per dump |
| 2 | refinement of the grid to sample in each direction (default is 1) |
| 3 | frequency of diagnosis (in dpd time units) |
| 4 | activeate the field dumps |
| 5 | frequency of field dumps (in dpd time units) |
The velocity profile is shown below:
